dist.matrix <- function(side)
{
row.coords <- rep(1:side, times=side)
col.coords <- rep(1:side, each=side)
row.col <- data.frame(row.coords, col.coords)
D <- dist(row.col, method="euclidean", diag=TRUE, upper=TRUE)
D <- as.matrix(D)
return(D)
}11 Autoregressive models
In general, autoregressive models should not be be fit with Jags, but with a
11.1 Example with Jags
This is based on code posted originally by Petr Keil http://www.petrkeil.com/?p=1910, to illustrate some comments I made in response to this blog post
11.1.1 Creating the data
This helper function that makes distance matrix for a side*side 2D array
Here is the function that simulates the autocorrelated 2D array with a given side, and with exponential decay given by lambda (the mean mu is constant over the array, it equals to global.mu)
cor.surface <- function(side, global.mu, lambda)
{
D <- dist.matrix(side)
# scaling the distance matrix by the exponential decay
SIGMA <- exp(-lambda*D)
mu <- rep(global.mu, times=side*side)
# sampling from the multivariate normal distribution
M <- matrix(nrow=side, ncol=side)
M[] <- rmvnorm(1, mu, SIGMA)
return(M)
}OK, finally simulating the data
# parameters (the truth) that I will want to recover by JAGS
side = 10
global.mu = 0
lambda = 0.3 # let's try something new
# simulating the main raster that I will analyze as data
M <- cor.surface(side = side, lambda = lambda, global.mu = global.mu)
image(M)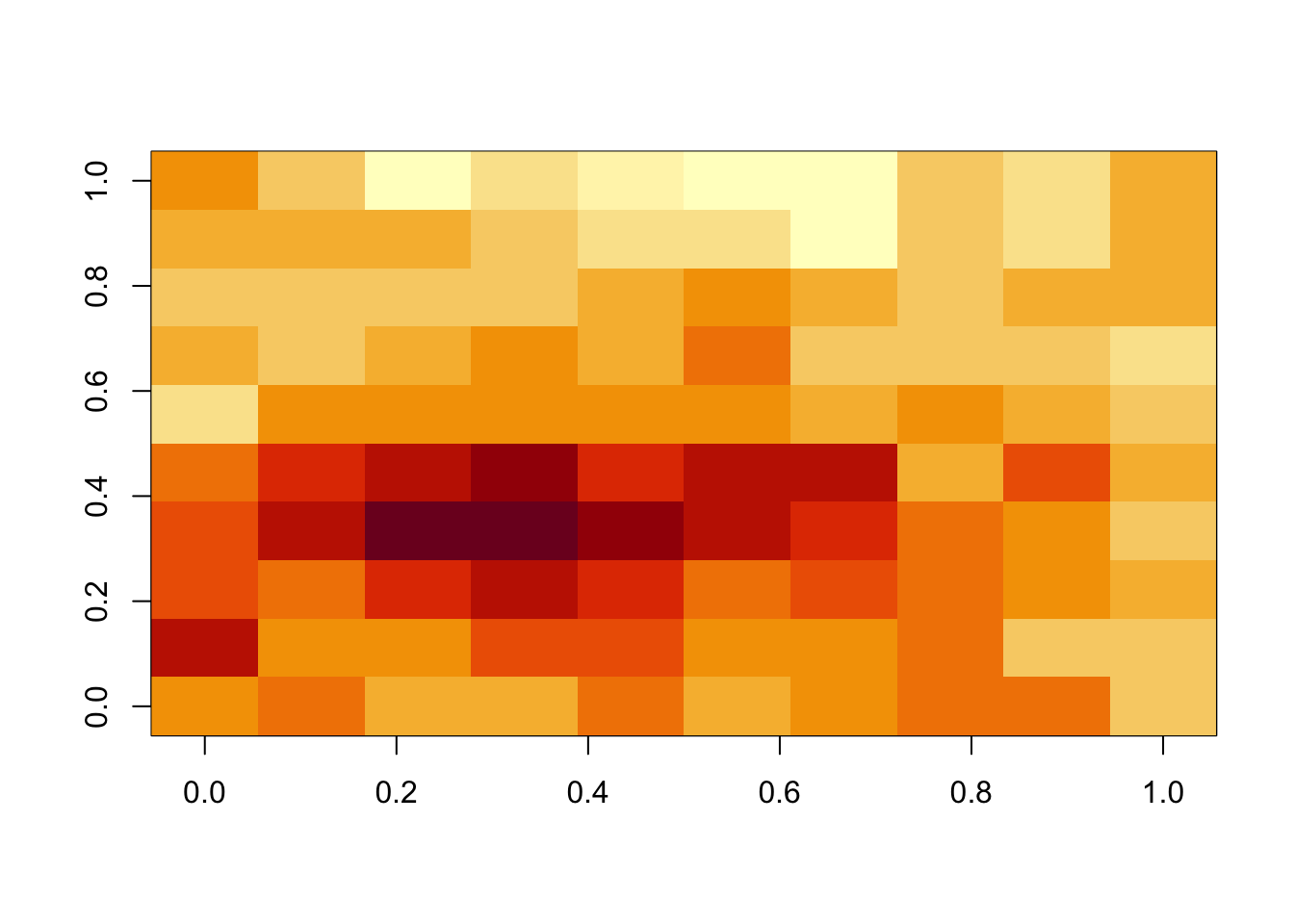
mean(M)[1] 0.4496296# simulating the inherent uncertainty of the mean of M:
#test = replicate(1000, mean(cor.surface(side = side, lambda = lambda, global.mu = global.mu)))
#hist(test, breaks = 40)11.1.2 Fitting the model in JAGS
preparing the data
y <- as.vector(as.matrix(M))
my.data <- list(N = side * side, D = dist.matrix(side), y = y)defining the model
modelCode = textConnection("
model
{
# priors
lambda ~ dgamma(1, 0.1)
global.mu ~ dnorm(0, 0.01)
for(i in 1:N)
{
# vector of mvnorm means mu
mu[i] <- global.mu
}
# derived quantities
for(i in 1:N)
{
for(j in 1:N)
{
# turning the distance matrix to covariance matrix
D.covar[i,j] <- exp(-lambda*D[i,j])
}
}
# turning covariances into precisions (that's how I understand it)
D.tau[1:N,1:N] <- inverse(D.covar[1:N,1:N])
# likelihood
y[1:N] ~ dmnorm(mu[], D.tau[,])
}
")Running the model
fit <- jags(data=my.data,
parameters.to.save=c("lambda", "global.mu"),
model.file=modelCode,
n.iter=10000,
n.chains=3,
n.burnin=5000,
n.thin=5,
DIC=FALSE)module glm loadedmodule dic loadedCompiling model graph
Resolving undeclared variables
Allocating nodes
Graph information:
Observed stochastic nodes: 1
Unobserved stochastic nodes: 2
Total graph size: 10114
Initializing modelplot(as.mcmc(fit))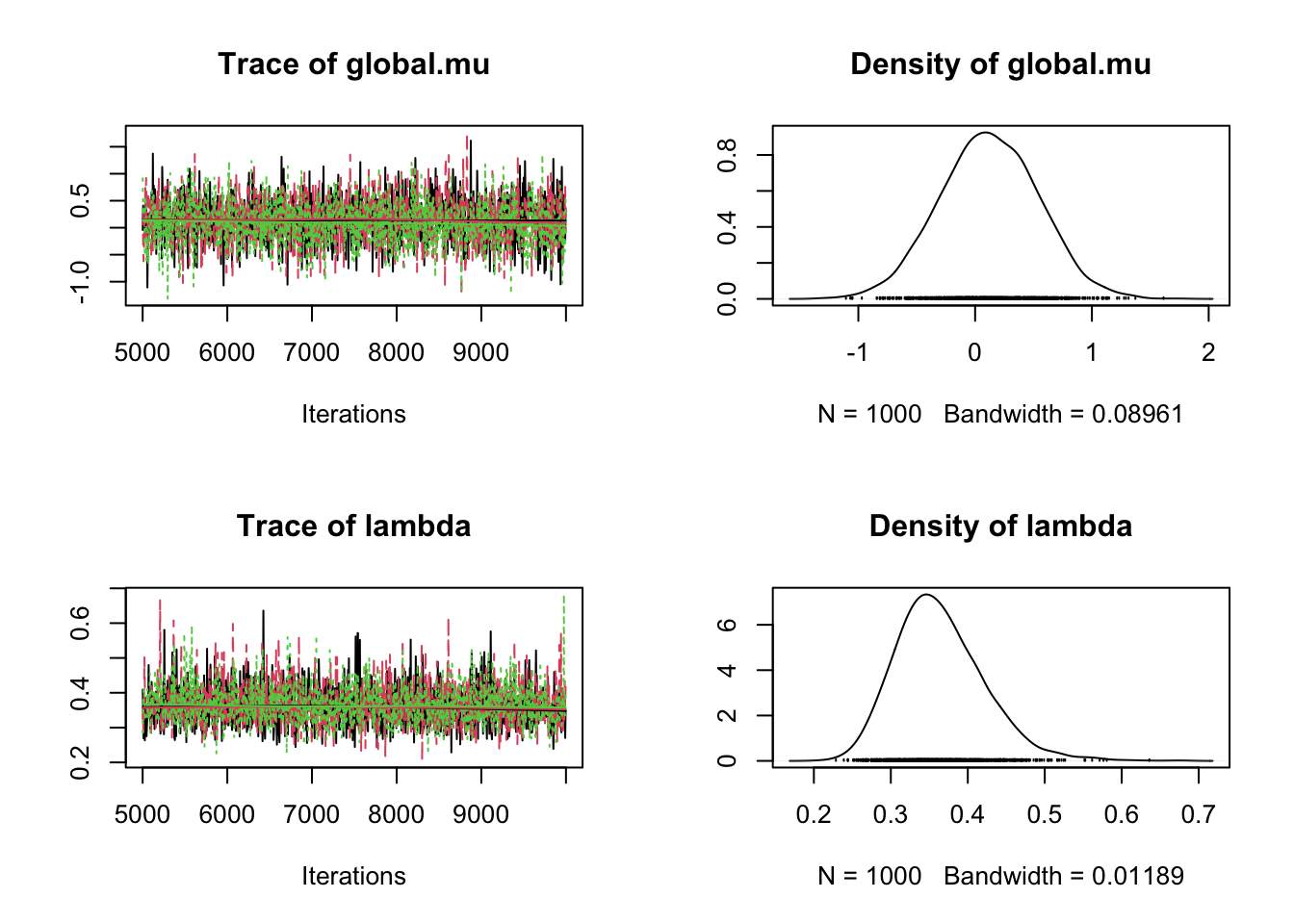
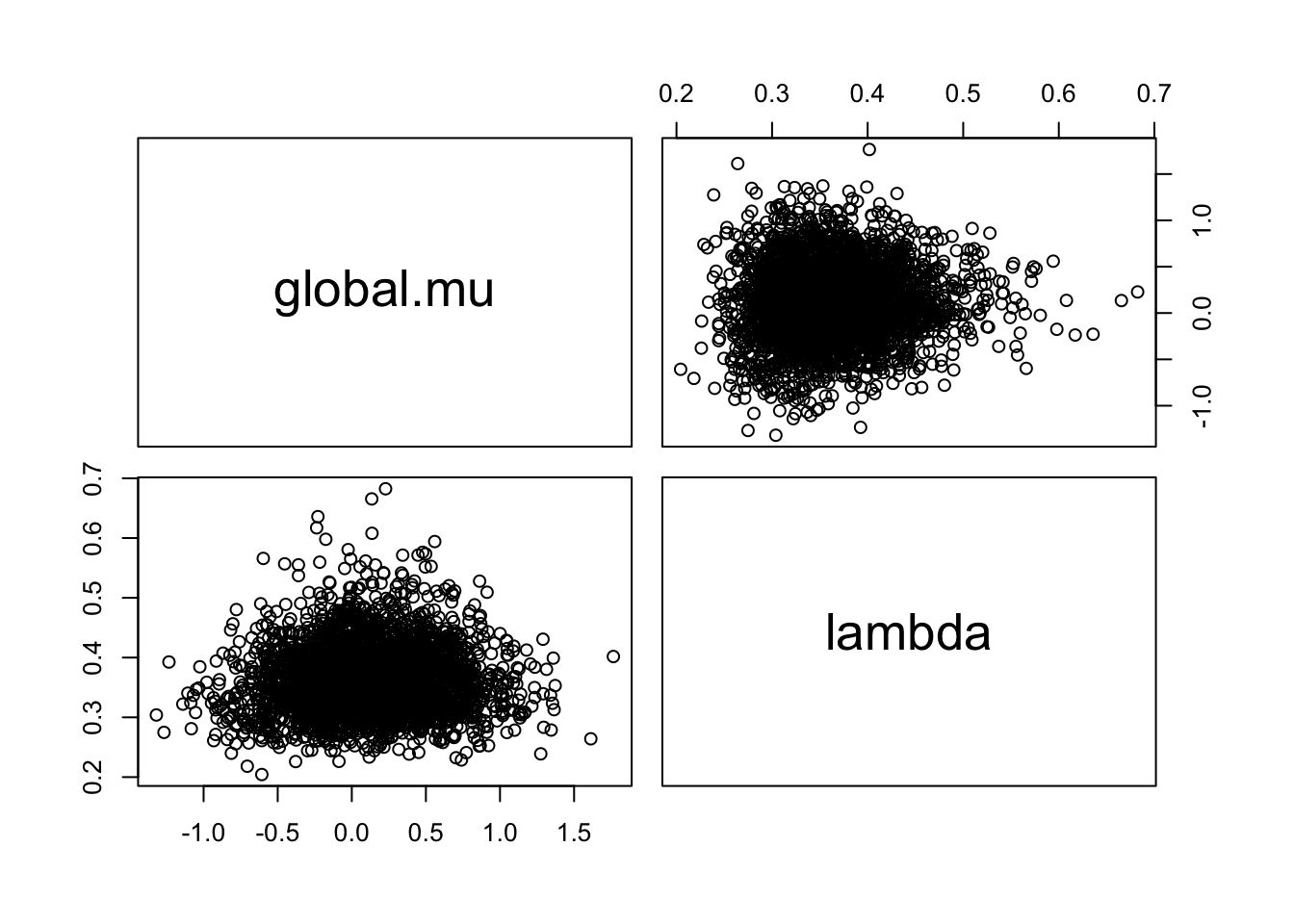
pairs(as.matrix(as.mcmc(fit)))